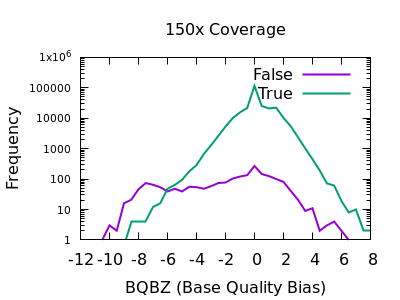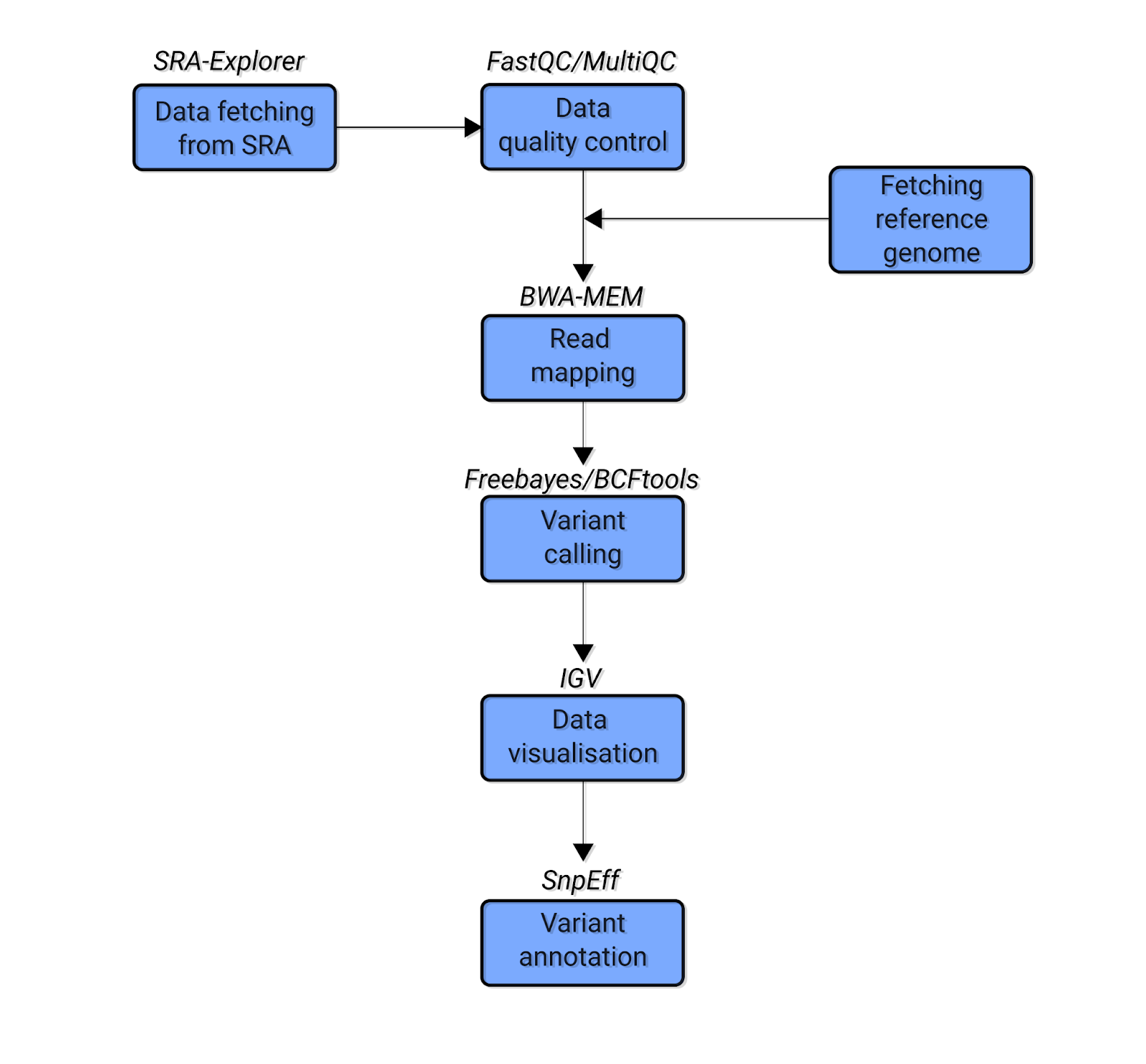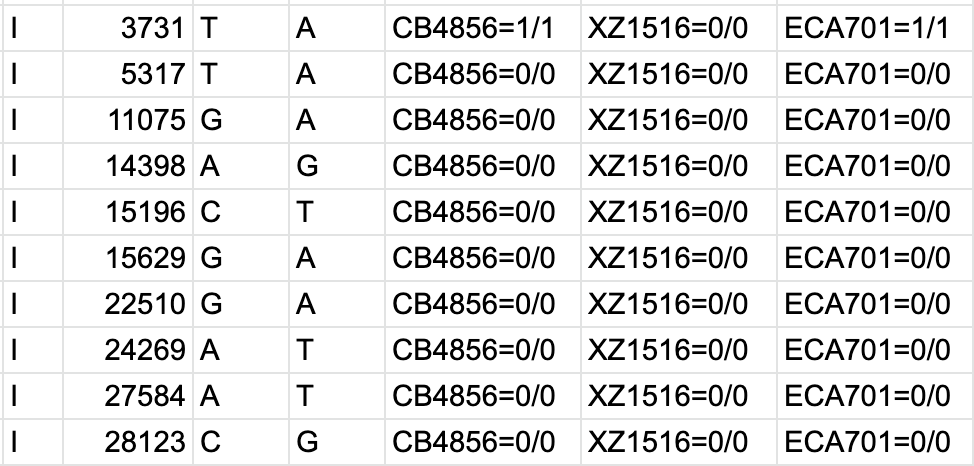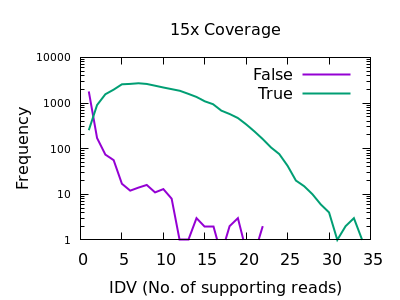
The evaluation of Bcftools mpileup and GATK HaplotypeCaller for variant calling in non-human species | Scientific Reports

Protocol for unbiased, consolidated variant calling from whole exome sequencing data - ScienceDirect

1744. Do you need to do Variant Quality Score Recalibration when calling somatic variants with Mutect2 - Legacy GATK Forum