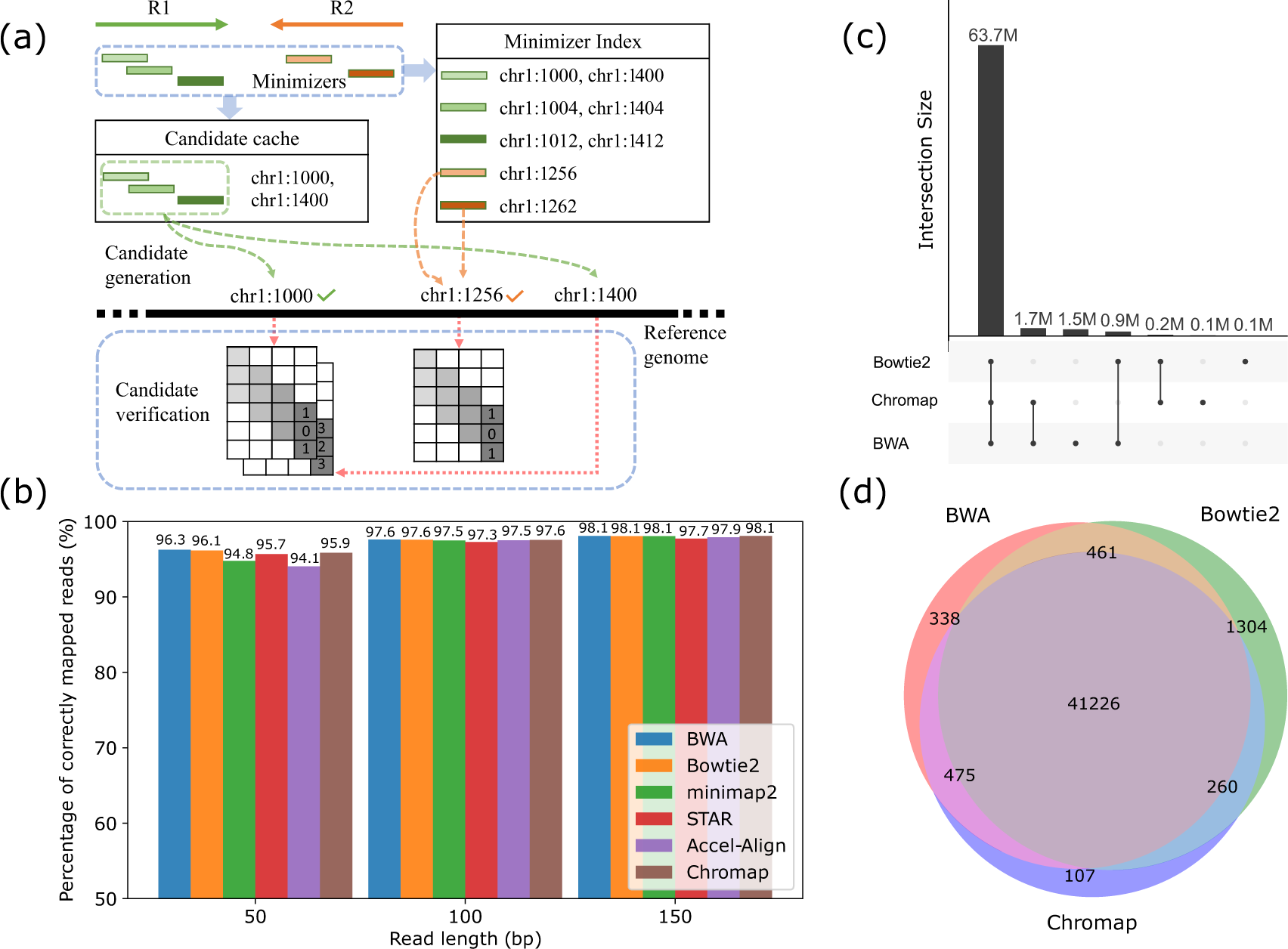
Seed extension by BWA-SW algorithm. Red substring 'TCCT' is the seed. A... | Download Scientific Diagram
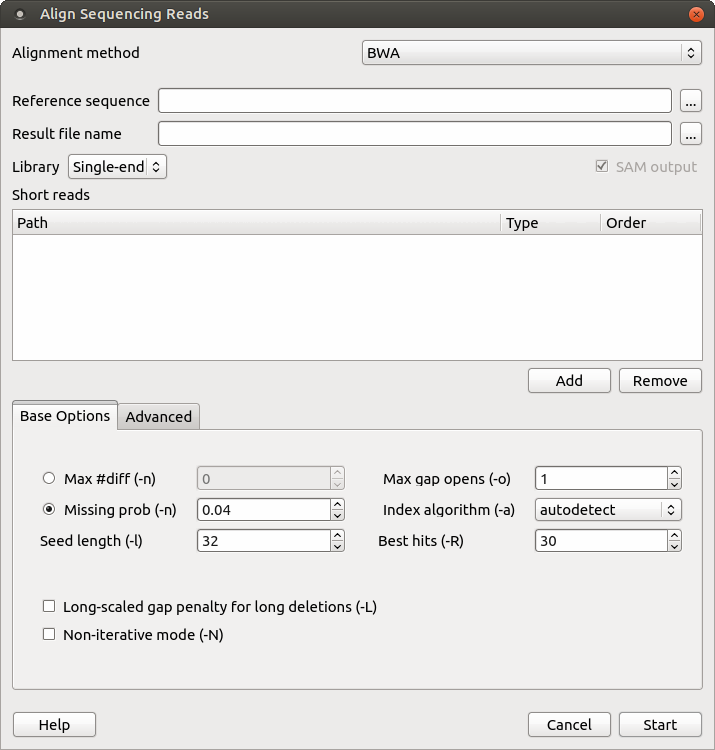
Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM : Heng Li : Free Download, Borrow, and Streaming : Internet Archive

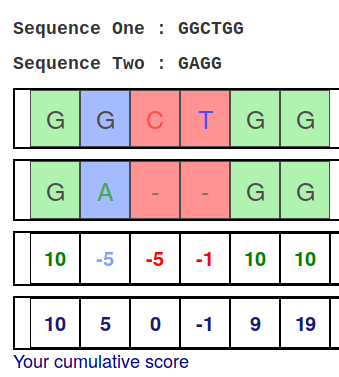
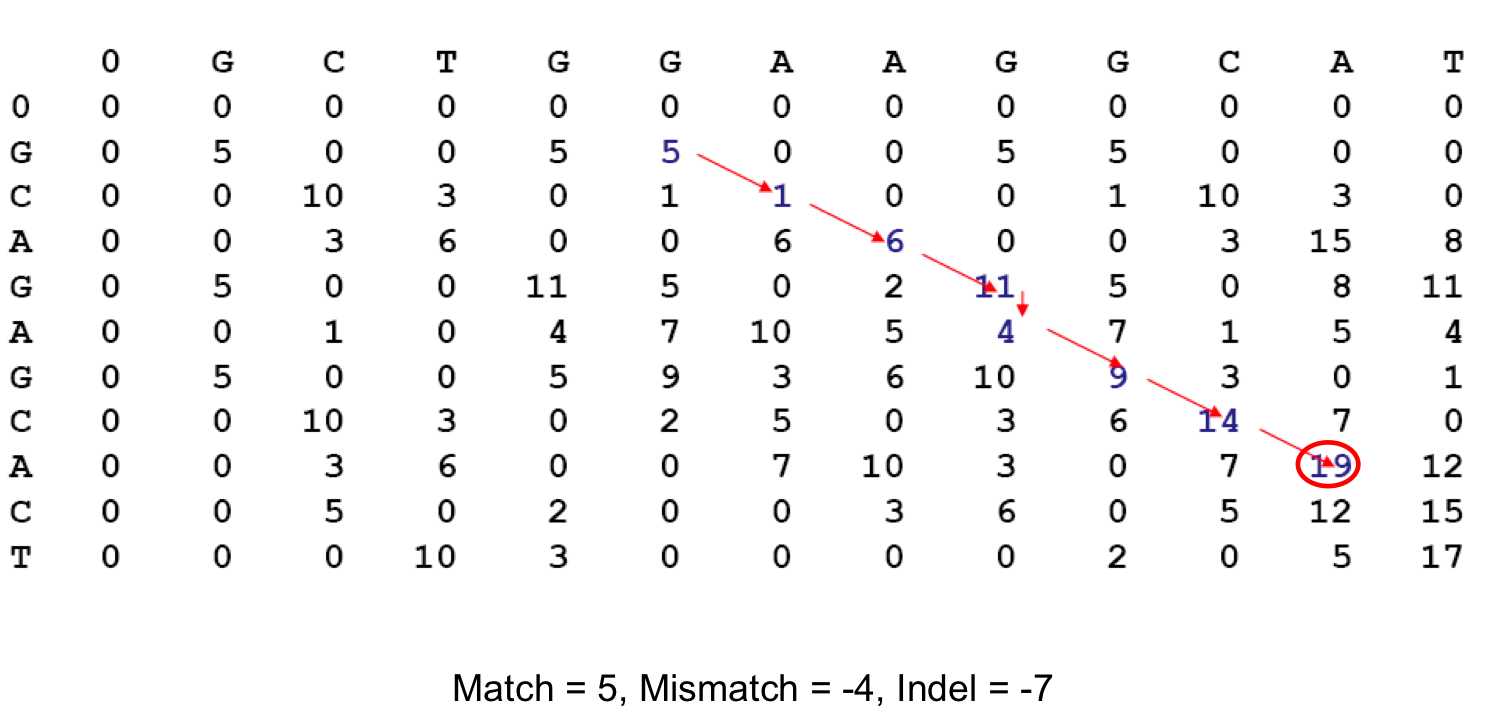
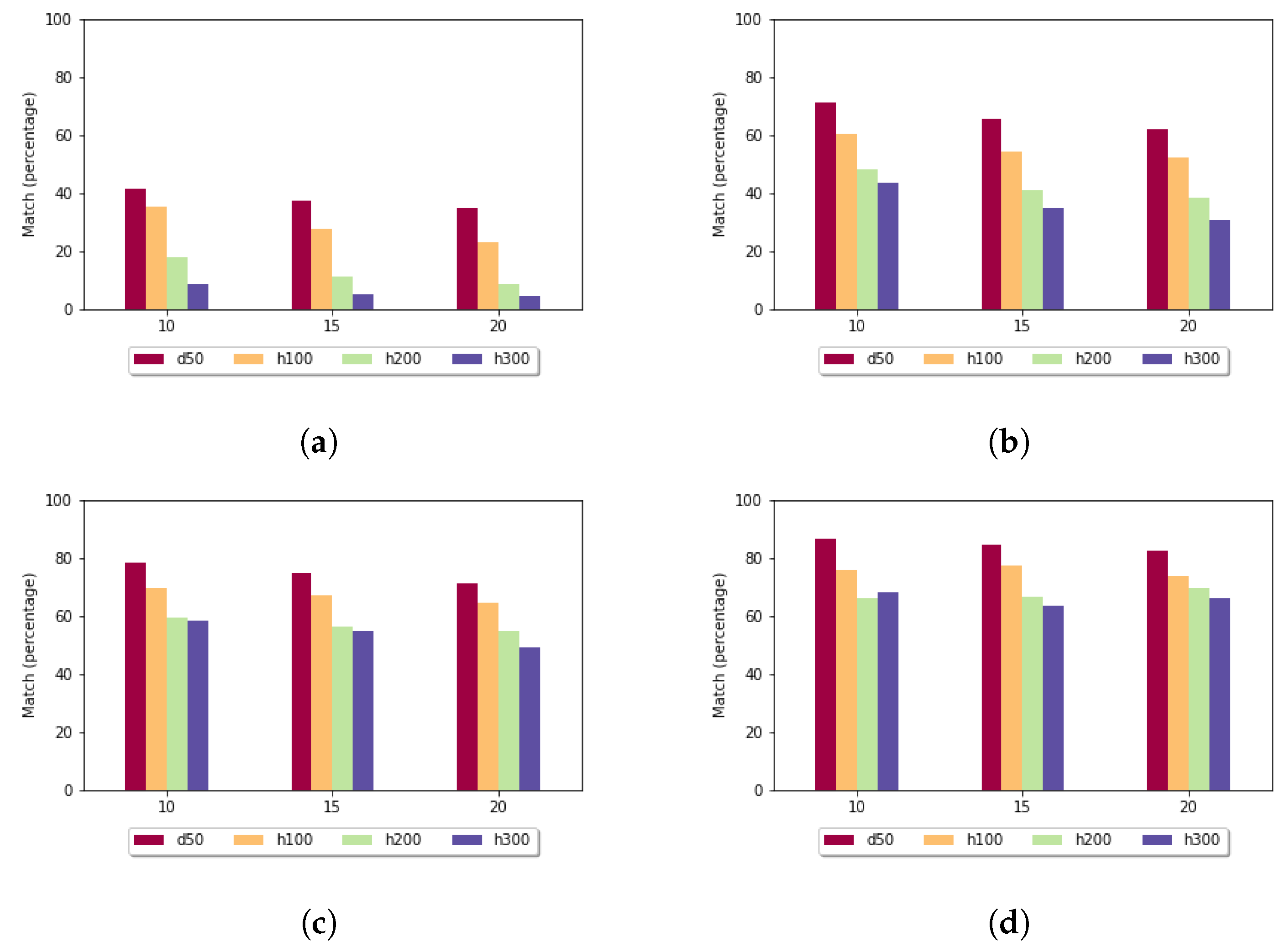
Pairwise alignment of nucleotide sequences using maximal exact matches | BMC Bioinformatics | Full Text

Pairwise alignment of nucleotide sequences using maximal exact matches | BMC Bioinformatics | Full Text

A high‐performance FPGA‐based BWA‐MEM DNA sequence alignment - Pham‐Quoc - 2021 - Concurrency and Computation: Practice and Experience - Wiley Online Library